Improving Animal Health and Welfare
Research into the health of individual animals is critical for ensuring their welfare, aids in conservation of wild populations, and provides insight into ecosystem health. We are developing innovative diagnostic methods such as MRIs, endoscopy and sonography, genome mapping of species, disentangling and rehabilitating individual animals. We are also developing cardiac and blood profiles for health assessments, and ecosystem screening for oncoming disease.
Projects and Recent Papers
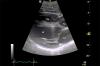
Assessing cardiac function in otariid seals as a diagnostic tool for assessing animal health (partner: NSERC)
2021 | ||||||||||||||
|
|

|
||||||||||||
2019 | ||||||||||||||

|
|

|
||||||||||||
2018 | ||||||||||||||

|
|

 |
||||||||||||
2015 | ||||||||||||||

|
|

 |
||||||||||||
