
New DNA technique offers dietary insights
Of all the species that share the North Pacific Ocean with humans, among the most visible are harbor seals. Some 40,000 harbor seals make their home in the Strait of Georgia, near Vancouver, and they prey on the same types of fish we do.

One of 40,000 harbor seals living in the Strait of Georgia.
Fisheries managers need to balance the dietary needs of these important predators with the needs of human fisheries. But how much fish do harbor seals need to consume in order to survive?
The precise answer to this question is notoriously elusive, but a new study led by Austen Thomas (The University of British Columbia) has provided scientists with a promising new technique to analyze both the composition and quantity of prey in a harbor seal’s diet.
“Up until now, the tools we have used to quantify the diets of seals have been inadequate for certain types of questions,” says Thomas. “They rely primarily on identifying the bones in the droppings of seals, and the resulting calculations produce only a rough estimate of what they’ve eaten. We’re using a tool that does a much better job of characterizing the prey species in a seal’s diet and—for the first time—gives us an idea of the quantity of each prey species present.”

Austen Thomas collecting harbor seal scats in the Fraser River estuary.
Sophisticated Technique
Thomas and colleagues have refined a promising new technique called DNA metabarcoding, which is used to identify specific marker genes in a sample of soil or water—or seal droppings. This sophisticated technique indicates whether the gene for a certain fish species is present or absent in a given sample, and its quantity relative to other species.
“We wanted to go further—to establish a link between the amount of prey DNA in a predator diet sample and the biomass of what they consumed,” Thomas says. “To do this, we prepared and analyzed fish samples of known biomass composition. This gave us a series of controls, which we compared against samples of seal droppings that contained an unknown prey composition.”

Vials of harbor seal scats soaking in ethanol to obtain prey DNA.
Thomas and colleagues developed a series of correction factors—a number unique to each fish species that helped to determine how much of each species-specific DNA marker occurred in the unknown samples.
Micro to Macro
After proving and replicating their technique, the researchers were able to establish an entire library of correction factors for different fish species. They then applied the correction factors to identify any interactions between species—for example, whether herring produced the same correction factor when mixed with pollock as it did when mixed with salmon. “The consistency of the individual correction factors across mixed samples was encouraging,” Thomas says.
“Our study aims to improve the overall accuracy of seal diet data,” says Thomas. “Small changes in how we measure an individual seal’s diet at the DNA level can lead to profoundly different estimates of fish consumed by the population. From a fisheries management perspective, it’s important to know what the 40,000 seals in the Straight of Georgia are eating because it has a potential impact on the things people like to consume. They’re an important predator.”
 PUBLICATION
PUBLICATION
|
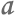
|

 |
||||||||||||
