
Researchers with their loot of scat
It might not be the most glamorous aspect of a biologist’s work, but the study of fecal matter is an invaluable (if messy) part of reconstructing an animal’s life history. Recently, however, Consortium researchers ushered this age-old research method into the 21st century by integrating DNA technology into the examination of animal scat.
The aim of the study was to determine the effects pinnipeds have on their prey populations — and by extension, on fisheries. In order to understand this, they needed to find out exactly what pinnipeds eat. Traditionally, researchers have reconstructed pinniped diets by probing scat samples for telltale treasures like fish skeletal remains (aptly known as “hard parts”), but this does not always account for the less-identifiable components of the scat’s “soft parts”. For example, if a predator only consumes the fleshy parts of the prey, skeletal remains will not be evident in the scat.
The researchers believed that DNA analysis of scat holds the key to unlocking the secrets of pinniped diets. They aimed not only to compose a comprehensive list of prey species, but also compare the effectiveness of this new technology to the old. The research team comprised Dominic Tollit and Andrew Trites of the UBC Fisheries Centre; Kristina Miller, Angela Schulze and Peter Olesiuk of the Pacific Biological Station; Susan Crockford of Pacific IDentifications; and Tom Gelatt and Rolf Ream of the Alaska Fisheries Science Centre. Their findings were recently published in Ecological Applications.

Wanted: Adventurous Fin-footed Eater
When it came to choosing a subject for the study, the researchers did not have to look far. With a penchant for fish and cephalopods, the Steller sea lion has a broad diet, and therefore is an excellent subject. Its diet has also been well studied since its western population numbers began to plummet in the 1980s. Plus, the Steller sea lion came with a mystery: though researchers had long understood its fondness for salmon, the salmon bones they recovered from the fecal samples largely failed to pinpoint the salmon species most important to its diet.
The research team collected scat samples in British Columbia and the eastern Aleutian Islands, and analyzed them through polymerase chain reaction (PCR) and denaturing gradient gel electrophoresis (DGGE) techniques. The DNA they extracted from the scat included that of over 40 different species of fish and cephalopods. Unexpectedly, they also found at least seven species of crustaceans such as spot prawns and Dungeness crab. Whether or not the sea lions had targeted the crustaceans or had consumed fish that had eaten the crustaceans is not known.
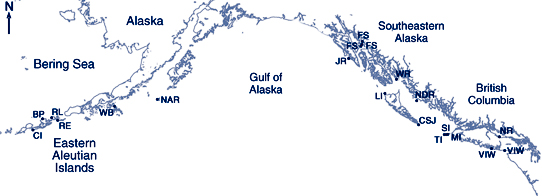
Steller sea lion scat collection site locations
Researchers using DNA analysis can now identify many more species and 20% more by number (occurrences) than they could using traditional methods of determining diets.
The researchers also solved one of the sea lion mysteries by finding that pink and chum followed by Chinook were the most important salmon prey. But in running the DNA analysis they also revealed a perplexing discovery when they found that two of the fifty-six scats collected in the Eastern Aleutian islands contained the DNA of Atlantic Salmon — presumably escapees that traveled to Alaska from salmon farms in British Columbia or further south.
Changing the Way We Think About Scat
The team concluded that DNA techniques can refine results not only in diet studies of pinnipeds whose scats lack hard parts, but also in diet studies of marine piscivores (fish-eaters) in general, including crustaceans, penguins, sea birds, and fish. DNA scat techniques have also been used to confirm the predator’s sex and the species and molecular techniques can determine hormones and parasites in scat. So while they might not raise scat science to the level of acceptable dinner conversation, they will surely continue to change the face of traditional research methods.
September 16, 2009
![]() RELATED PUBLICATION:
RELATED PUBLICATION:
|
|

 |
||||||||||||
