
New Model May Help to Quantify Prey Consumption
Harbor seals play an important predatory role in the North Pacific Ocean, and the types of fish they prefer to eat are well known. But scientists know little about the relative proportion of those fish in the diet (e.g., how much salmon is eaten compared to herring?). This knowledge would be useful to fisheries managers, for example, in projecting the impact of Harbor seals on various fish stocks.
A new research technique has put scientists one step closer to quantifying the diet of Harbor seals and other marine mammals. Rather than relying on imperfect procedures, such as extracting fish bones from feces or stomach contents, scientists can analyze the chemical remains of prey species contained in the blubber to determine their relative proportions in the diet.
Consortium researchers recently tested an analytic method, termed Quantitative Fatty Acid Signature Analysis (QFASA), on a group of Harbor seals. The study, authored by Chad Nordstrom, Lindsay Wilson, Dominic Tollit (each of the University of British Columbia) and Sara Iverson (Dalhousie University), was recently published in Marine Ecology Progress Series.
Quantifying Fatty Acids
The study was conducted over a six-week period and involved 21 Harbor seals temporarily housed at the Vancouver Marine Mammal Rescue facility. Each seal was estimated to be less than a month old, representing a dietary “clean slate.” The seals received one of three diets: smelt only, herring only, or three weeks of smelt and three weeks of herring. A blubber biopsy was taken from each animal at the beginning, middle, and end of the study.
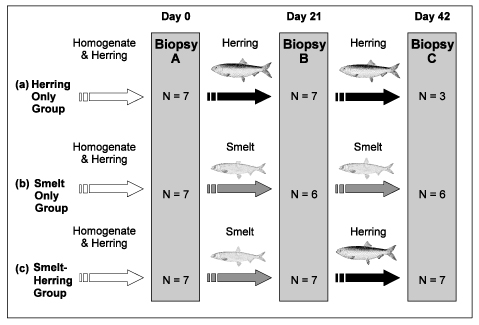
Fig. 1. Feeding regimes for (a) Herring Only, (b) Smelt Only, and (c) Smelt-Herring treatment groups over 42 days. Number of seals (N) sampled at each biopsy noted for each group. Coloured arrows indicate diet fed (white = homogenate & herring, black = herring, grey = smelt).
Under these controlled circumstances, the researchers sought to determine whether the QFASA model would correctly identify the known, six-week dietary regimes. QFASA analysis is based on the principle that long-chain fatty acids from prey species, each of which have a unique and identifiable chemical “signature”, are predictably incorporated into the predator’s blubber. Once the predator’s own fatty acid signature is obtained from the biopsy sample, it can be compared with a library of established fatty acid signatures from potential prey.
However, not all of the 68 fatty acids that may be present in Harbor seal blubber are obtained from their diet; some may be created by the seals through metabolism. Thus the researchers proposed to use a “reduced subset” of 35 fatty acids that appear most strongly associated with diet. Further, to correct for any changes to the prey’s fatty acid signature that may occur during digestion, the researchers obtained a species-specific calibration coefficient. The calibration coefficient and the fatty acid subset are two important inputs into the QFASA model that could significantly influence the results. Thus the researchers also sought to assess the sensitivity of the model to these two key inputs.
Promising Model
The study showed that over 42 days of captive feeding, the QFASA model could directly distinguish which seals were on single-species diet. However, results were inconclusive for the seals on mixed diets. The researchers found that the reliability of the results was most strongly influenced by the calibration coefficient and the fatty acid subset that were input into the model.
Thus the QFASA model shows promise in eventually helping researchers to evaluate the quantitative composition of predator’s diets. The current study demonstrated that the model is effective in assessing a constant diet, but further research is needed to effectively quantify mixed diets, especially those containing an unknown array of prey species. QFASA may prove useful in research on wild marine mammal populations once its effectiveness has been established though further captive studies.
July 21, 2008
![]() PUBLICATION
PUBLICATION
|
|

 |
||||||||||||
